ASaiM
Lessons learned from developing a framework for biologistsBérénice Batut — October 16th, 2015

PhD thesis in bioinformatics and computational biology
|
Post-doc in bioinformatics
|

Gut microbiota
Community of microorganism species that live in the digestive tracts
"Forgotten" organMetagenomic: study of microbiota
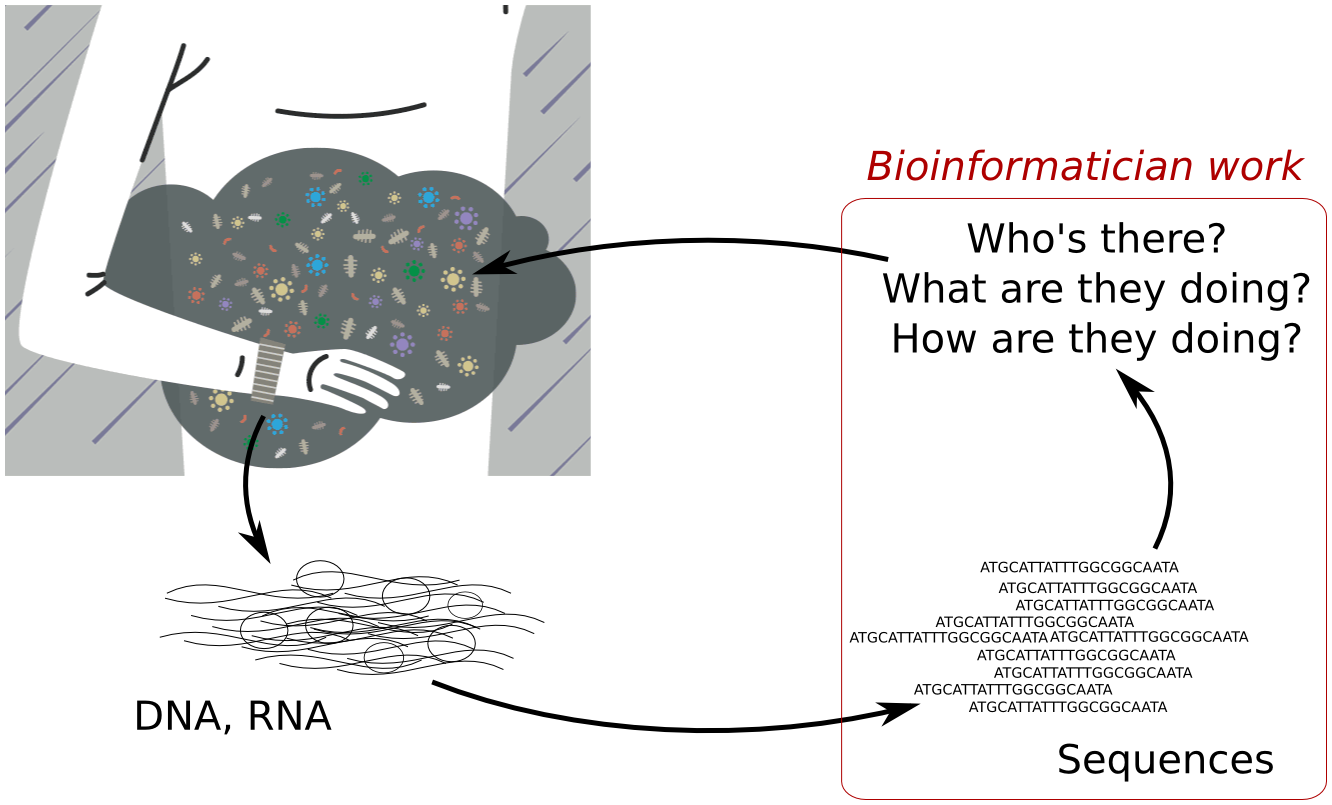
Example of workflow to sort sequences given their type